
Fuzzy C-Means Clustering: Comprehensive Guide 2024
FCM clustering lets data points belong to multiple groups at once, unlike traditional clustering that forces points into single groups.
Here's what makes FCM different from other clustering methods:
| Feature | FCM | Traditional Clustering |
|---|---|---|
| Point Assignment | Can be in multiple clusters (0-100%) | Only one cluster (100%) |
| Noise Handling | Works better with messy data | Gets thrown off by outliers |
| Speed | Takes longer to process | Faster but less detailed |
| Best Use Cases | Medical imaging, gene analysis, pattern finding | Simple data grouping |
Key Things to Know:
- Each data point gets percentage scores for different clusters
- Works great with overlapping groups and messy data
- Takes more processing time but handles real-world data better
- Perfect for medical imaging, gene studies, and finding hidden patterns
Common Problems and Solutions:
- Starting points matter - bad starts = wrong results
- Picking cluster numbers is tricky - use testing methods
- Gets slow with big data - batch processing helps
Real-World Uses:
- Medical: Groups patient data and analyzes images
- Biology: Maps gene patterns and protein interactions
- Business: Finds customer segments and market trends
- Research: Sorts documents and finds topic connections
Want to use FCM? Start with clean data, pick your cluster count carefully, and be ready to trade some speed for accuracy.
Related video from YouTube
FCM Basics
Let's break down how FCM works (and how it's different from K-Means).
FCM vs K-Means
Here's what makes FCM special:
| Feature | FCM | K-Means |
|---|---|---|
| Data Point Assignment | Points can belong to multiple clusters (0-100%) | Points belong to only one cluster (100%) |
| Noise Handling | Less affected by outliers | More sensitive to outliers |
| Processing Speed | Slower due to extra calculations | Faster processing |
| Cluster Shapes | Works with various shapes | Best with spherical shapes |
| Data Overlap | Handles overlapping data well | Struggles with overlap |
Math Behind FCM
Let's look at the two key formulas that power FCM:
1. Objective Function
FCM uses this formula to find the best clusters:
J = Σ(i=1 to n) Σ(j=1 to c) wij^m · d^2(xi, vj)
Here's what each part means:
- n = number of data points
- c = number of clusters
- wij = membership value
- m = fuzziness parameter (>1)
2. Membership Updates
This formula shows how FCM updates point memberships:
wij = 1 / Σ(k=1 to c)(d(xi,vj)/d(xi,vk))^(2/(m-1))
Main FCM Parts
Here are the key pieces that make FCM work:
| Component | Purpose | Details |
|---|---|---|
| Membership Matrix | Tracks cluster belonging | Values between 0-100% |
| Cluster Centers | Define group locations | Updated each iteration |
| Fuzziness Parameter | Controls overlap amount | Usually set between 1.5-3.0 |
| Distance Measure | Calculates point spacing | Often Euclidean distance |
| Stopping Criteria | Determines completion | Based on change threshold |
FCM follows these steps:
- Pick random cluster centers
- Calculate how much each point belongs to each cluster
- Move cluster centers based on these calculations
- Keep going until the changes get tiny
To use FCM, you'll need to set:
- Number of clusters (c)
- Fuzziness parameter (m)
- Maximum iterations
- Stopping threshold
How FCM Works
FCM splits data into clusters through a probability-based approach. Let me break down how it works.
FCM Structure
The algorithm uses 4 main parts to get the job done:
| Component | Description | Purpose |
|---|---|---|
| Membership Matrix | Values from 0-1 | Shows each point's cluster membership strength |
| Objective Function | J = Σ(i=1 to n) Σ(j=1 to c) wij^m · d^2(xi, vj) | Finds optimal cluster assignments |
| Convergence Check | Tracks iteration changes | Tells algorithm when to stop |
| Distance Calculator | Measures point-center gaps | Helps set membership values |
Getting Started with FCM
Here's how FCM works, step by step:
1. Set Up Your Starting Point
First, you need to:
- Pick how many clusters you want (c)
- Set random starting points for cluster centers
- Choose your fuzziness setting (m > 1)
2. Work Out Memberships
For each piece of data:
- Calculate how far it is from each cluster center
- Use this formula to figure out membership values:
wij = 1 / Σ(k=1 to c)(d(xi,vj)/d(xi,vk))^(2/(m-1))
3. Move the Centers
Shift cluster centers based on where your data points are (using weighted averages).
Making FCM Better
Tests show these tweaks can make FCM work better:
| Change | Result | How to Do It |
|---|---|---|
| Weight Parameters | 25% faster | Add weights to membership math |
| Modified Distance | Handles messy data better | Use weighted distances |
| Adaptive Fuzziness | More precise clusters | Change m as you go |
Looking at tests with dataset X12:
- Basic FCM: Takes 12 rounds
- FCM with weights: Only 9 rounds
- Closest to actual center: 0.1537
The Weight Possibilistic FCM (WPFCM) version:
- Handles messy data better
- Gets results faster
- Finds cluster centers more accurately
Making FCM Better
Let's look at how to get better results from FCM clustering.
Picking the Right Settings
The fuzziness parameter (m) is KEY for FCM performance. Data shows you'll get the best results with m values between 1.5 and 2.5.
Here's what matters most:
| Parameter | Optimal Range | Impact on Results |
|---|---|---|
| Fuzzifier (m) | 1.5 - 2.5 | Controls noise tolerance |
| Cluster Size | Based on data | Affects minority clusters |
| Feature Weights | 0 - 1 | Shows feature importance |
Speed and Accuracy Tips
Tests on UCI datasets point to some clear winners for better FCM:
| Method | Speed Gain | Accuracy Boost |
|---|---|---|
| EFMC Algorithm | 2.33x faster | 98.3% at epoch 30 |
| vFCM Method | Less tuning needed | Similar to k-means |
| PCA + Combined Distance | 15 iterations | 1.6468 cluster accuracy |
The EFMC method CRUSHES the competition:
- Makes loss values 2.71x better
- Boosts accuracy by 2.5x
- Runs 2.05x faster when data is 50% homogeneous
Working with Big Data
When your dataset gets huge, here's what works:
| Technique | Purpose | Result |
|---|---|---|
| PCA Reduction | Cut dimensions | More stable clusters |
| Minkowski-Chebyshev | Better similarity measure | 0.0373 objective value |
| Genetic Algorithm | Parameter optimization | Better cluster numbers |
Check out these improvements on the glass dataset:
| Metric | Standard FCM | Better FCM |
|---|---|---|
| F-value | 0.7843 | 0.8302 |
| G-mean | 0.8552 | 0.8970 |
| Accuracy | 0.8972 | 0.9159 |
Size-aware FCM beats basic FCM for uneven groups:
| Dataset | Accuracy Increase |
|---|---|
| Wine | +3.93% |
| Glass | +1.87% |
| User Knowledge | +19.98% |
Dealing with Bad Data
Bad data messes up FCM in two main ways:
| Issue Type | Impact on FCM | Detection Method |
|---|---|---|
| Noise | Reduces accuracy by 23-45% | Check membership values entropy |
| Outliers | Skews cluster centers | Monitor distance from centroids |
| Missing Values | Creates false patterns | Data completeness analysis |
Here's what works better than standard FCM when your data's messy:
| FCM Version | Best For | Performance Boost |
|---|---|---|
| FCM_S1/S2 | Image noise | +15% accuracy |
| FGFCM | Mixed noise types | 2x faster convergence |
| HMRF_FCM | Local patterns | +27% noise resistance |
| FLICM | Spatial data | 3x better with outliers |
Let's look at fixes that actually work:
| Problem | Solution | Results |
|---|---|---|
| Image Noise | Use local spatial info | 98% noise reduction |
| Measurement Noise | Apply k-means pre-filtering | +31% accuracy |
| Mixed Data Types | Two-stage clustering | 87% correct grouping |
NASA's software projects showed this simple process works:
- Cut out the 5% noisiest points
- Look at membership values
- Run FCM again on clean data
This brought error rates DOWN from 12% to 3.8% in ultrasonic sensor data.
Want better results? Do this:
- Look at data quality first
- Match FCM type to your noise
- Use nearby data points for spatial stuff
- Get rid of obvious outliers
- Compare results with known patterns
Here's proof it works: In MRI brain scans, NR-IFCM beat basic FCM by cutting noise impact by 76%. How? By mixing:
- Local gray-level data
- Spatial patterns
- Membership linking
Using Python for FCM
Here's how to implement FCM in Python:
from __future__ import division, print_function
import numpy as np
import matplotlib.pyplot as plt
import skfuzzy as fuzz
# Make test data
centers = [[4, 2], [1, 7], [5, 6]]
sigmas = [[0.8, 0.3], [0.3, 0.5], [1.1, 0.7]]
np.random.seed(42)
The main tools you'll need:
| Component | What It Does | Output |
|---|---|---|
| skfuzzy.cmeans | Handles clustering | Membership matrix |
| numpy arrays | Prepares data | Clean data format |
| cmeans() | Does clustering | Group memberships |
| cmeans_predict() | Labels new data | Classifications |
Setting Up FCM
These settings control how FCM works:
| Setting | What It Does | Typical Range |
|---|---|---|
| n_clusters | Sets group count | 2-10 |
| max_iter | Sets max cycles | 100-1000 |
| error | Sets stop point | 0.005-0.01 |
| random_state | Makes results match | 42 |
Here's the basic code:
fcm = FCM(n_clusters=3)
fcm.fit(X)
fcm_labels = fcm.u.argmax(axis=1)
Making Code Run Better
Want faster code? Try these:
| Change | Speed Gain | Memory Impact |
|---|---|---|
| NumPy arrays | 4x faster | No change |
| Pre-filtering | 2x faster | 30% less |
| Batch processing | 3x faster | 20% more |
Here's a simple example:
# Quick FCM setup
from fcmeans import FCM
X, _ = make_blobs(n_samples=50000, centers=[(-5, -5), (0, 0), (5, 5)])
fcm = FCM(n_clusters=3)
fcm.fit(X)
Check how well it worked:
print(f"FPC: {fcm.fpc}") # 0 to 1 scale
Want good clusters? Look for FPC scores above 0.7.
sbb-itb-4f108ae
Common FCM Problems
FCM clustering has 3 main problems that can mess up your results. Here's what you need to know:
Starting Point Problems
Your starting point makes a BIG difference in FCM. Bad starts = bad results.
| Problem | What Happens | How to Fix |
|---|---|---|
| Gets stuck in local spots | Wrong clusters | Run FCM several times |
| Results keep changing | Different answers each time | Start with K-means centers |
| Takes too long | Wastes processing time | Start with spread-out points |
Here's what works best: Use K-means first, THEN run FCM. It takes extra time but stops those bad starts.
Picking Cluster Numbers
You need the right number of clusters. These methods help:
| Method | What It Does | Best Use Case |
|---|---|---|
| Elbow method | Shows errors vs clusters | Smaller datasets |
| Silhouette check | Shows how well separated | Mixed clusters |
| Quality index | Tests cluster quality | Big datasets |
Test different numbers and use these methods to check what works best.
Speed Issues
FCM gets SLOW with big data. Here's the breakdown:
| Problem | Time Cost | Fix |
|---|---|---|
| Too many dimensions | Gets complex fast | Cut dimensions first |
| Loops too much | Can hit 1000 cycles | Use 0.73 threshold |
| Too much data | Slow processing | Process in batches |
Pro tip: Set your threshold to 0.73. You'll cut processing time by 75.2% and only lose 2% quality.
The time math looks like this: O(ndc²t)
- n = data points
- d = dimensions
- c = clusters
- t = loops
To make it faster:
- Clean your data first
- Process in chunks
- Set smart limits
- Start in good spots
Bottom line: You'll need to pick between speed and perfect results.
Where to Use FCM
FCM works best in three key areas:
Finding Patterns
FCM spots patterns in data that humans might miss. Here's what it can do:
| Data Type | Use Case | Results |
|---|---|---|
| Gene Expression | Protein interaction analysis | Groups similar genes |
| Time Series | Market trend analysis | Shows buying patterns |
| Customer Data | Behavior segmentation | Maps shopping habits |
Take E. coli studies: FCM groups similar metabolic responses, making it easier to understand how these organisms work.
Working with Images
FCM breaks down medical images with high accuracy:
| Application | What FCM Does | Success Rate |
|---|---|---|
| MRI Scans | Spots brain tumors | Better than standard methods |
| Mammograms | Finds breast lesions | Speeds up detection |
| Medical Imaging | Segments tissue types | Cuts review time |
Back in 2011, doctors used FCM to find early-stage breast cancer in mammograms - and it worked FASTER than manual checks.
Biology Data
FCM handles complex bio data like a pro:
| Field | Application | Key Benefit |
|---|---|---|
| Gene Analysis | Groups similar expressions | Maps gene connections |
| Disease Typing | Clusters patient data | Improves treatment plans |
| Drug Testing | Tracks metabolic changes | Makes research faster |
"FCM clustering makes feature extraction simple by splitting different attributes into clusters - that's KEY for getting medical imaging right."
It's perfect for:
- Protein interactions
- Metabolic pathways
- Disease patterns
- Treatment responses
For teams using Zemith's AI document analysis, FCM processes big datasets and finds biological patterns WAY faster than manual work.
FCM with Modern Tools
Let's look at how today's tools make FCM more powerful and easier to use.
AI Tools and FCM
AI platforms take FCM to the next level. Here's what the top tools can do:
| Platform | FCM Features | Main Use |
|---|---|---|
| SageMaker | Built-in FCM support | Large dataset clustering |
| RapidMiner | GUI for FCM workflows | Visual data analysis |
| DataRobot | Automated FCM models | Predictive analytics |
| IBM Watson | FCM integration | Pattern detection |
Teams using Zemith's document analysis get THREE big benefits:
- Quick topic clustering
- Content relationship mapping
- Research paper grouping
Document Analysis
Want to sort documents FAST? FCM does the heavy lifting:
| Task Type | How FCM Helps | Results |
|---|---|---|
| Text Classification | Groups similar content | Sorts by topic |
| Citation Analysis | Links related papers | Shows research connections |
| Content Organization | Clusters documents | Creates topic maps |
The fclust package (2.1.1) comes with:
-
Fclustfor quick setup - Smart cluster selection
- Ways to see your results
Research Tools
Here's how FCM connects with research software:
| Tool | Integration | Output |
|---|---|---|
| KNIME | 300+ data connectors | Machine learning models |
| MonkeyLearn | Text analysis focus | Content clusters |
| Power BI | Data visualization | Interactive dashboards |
MetaCluster pairs FCM with meta-learning to pick the best clustering method. PandasAI brings FCM to Python's data tools - no fancy coding needed.
"The package includes fuzzy clustering algorithms for both object data and relational data, allowing for a wide range of applications in various fields."
Need proof it works? The Galaxy Zoo project used FCM to sort galaxies based on multiple people's observations. That's the kind of complex work FCM handles every day.
Testing FCM Results
FCM testing needs clear metrics to measure performance. Let's look at the main ways to check if your FCM is working right.
Test Methods
Here are the 3 key metrics you'll want to track:
| Metric | What It Measures | Target Score |
|---|---|---|
| Silhouette Score | How well data points fit their clusters | 0.5+ |
| Davies-Bouldin Index | Cluster separation vs. spread | Below 1.0 |
| Adjusted Rand Index (ARI) | Match with known groupings | Above 0.7 |
Don't just pick one metric and call it a day. Data from six microarray tests shows that combining these measures gives you a much better picture of how well your clustering works.
Checking Results
Here's what the numbers should look like when you test:
| Step | Action | Expected Output |
|---|---|---|
| Data Matrix | Create K x S comparison | Cluster vs. actual classes |
| Precision Check | Calculate correct assignments | Target: >85% |
| Recall Analysis | Measure found vs. total items | Target: >85% |
| F1-Score | Combined precision/recall | Target: >87% |
These aren't just random targets. A 5-cluster test backed them up:
- Precision hit 91.50%
- Recall reached 87.94%
- F1-score landed at 89.68%
FCM vs Other Methods
Let's compare FCM with its main competitor:
| Feature | FCM | K-Means |
|---|---|---|
| Speed | Slower due to calculations | Faster processing |
| Accuracy | Better for overlapping data | Better for clear divisions |
| Memory Use | Higher | Lower |
For big datasets, tools like Zemith's document analysis can speed things up. Their AI handles complex clustering fast, which helps a lot with text and research data.
FCM shines with biological data too. Testing on the Yeast II microarray dataset, FCM found gene groups with p-values of 6.09E-16 - way better than standard clustering methods.
What's Next for FCM
FCM keeps getting better. Here's what's happening:
New Ideas
The HPFCM method is changing the game. Check out these numbers:
| Improvement | Speed Gain | Quality Gain |
|---|---|---|
| SPAM Dataset | 97.65% fewer iterations | 82.42% better results |
| ABALONE Dataset | 98.17% fewer iterations | 5.67% quality loss |
A new way to speed things up uses thresholds:
| Threshold | Iteration Reduction | Quality Impact |
|---|---|---|
| 0.73 | 75.2% decrease | 2% quality loss |
| 0.35 | 64.56% decrease | 1% quality loss |
Study Topics
FCM is booming. In 2021, researchers published 1,282 papers in Web of Science Core Collection. Here's where the action is:
| Research Area | Current Status | Next Steps |
|---|---|---|
| Image Processing | Most active field | Pixel grouping improvements |
| Big Data | Growing applications | Speed optimization for large sets |
| Natural Language | New development area | Text clustering methods |
FCM and AI
FCM + AI = Better Results. Here's how they work together:
| Integration Type | Purpose | Results |
|---|---|---|
| Explainable AI | Better understanding | Clear cluster interpretations |
| Deep Learning | Pattern recognition | More accurate grouping |
| Automated Tools | Faster processing | Reduced manual work |
Zemith's tools make FCM text processing FAST. And the new RL-MFCM algorithm? It's a game-changer:
- Starts working right away
- Finds better cluster centers
- Handles any kind of data
"This study shows that FCM has great potential in bibliometric analysis, especially in classifying and identifying the main topics of scientific publications." - Samsul Arifin
Summary
FCM does things differently than other clustering methods. Here's what makes it stand out:
| Feature | FCM | Traditional Clustering |
|---|---|---|
| Data Point Assignment | Multiple clusters (0-1 range) | Single cluster only |
| Noise Handling | Less affected by outliers | More sensitive |
| Processing Speed | More calculations needed | Faster processing |
| Accuracy | Higher for overlapping data | Better for distinct groups |
Let's look at how FCM performs against K-means:
| Dataset Type | FCM Accuracy | K-Means Accuracy |
|---|---|---|
| Liver Disorders | 52.79% | 55.43% |
| Wine Data | 68.54% | 70.22% |
| Class 1 Data | 11.97% | 9.85% |
| Class 2 Data | 81.91% | 87.94% |
Want to get the most out of FCM? Here's what works:
- Run it multiple times: Different starting points = better results
- Pick the right settings: Your fuzziness value (m) matters
- Compare your results: Check against other clustering methods
| Action | Why Do It | Result |
|---|---|---|
| Multiple Runs | Cuts down random variation | Better cluster centers |
| Parameter Testing | Fits your data structure | More accurate groups |
| Result Validation | Backs up your findings | Higher confidence |
FCM works especially well for:
- Image processing
- Finding patterns in complex data
- Biological data analysis
- Marketing segments
Here's the bottom line: If your data points might fit in multiple groups, FCM is your friend. But if you're dealing with clear-cut categories, you might want to keep it simple and use other methods.
Related posts
Explore Zemith Features
Introducing Zemith
The best tools in one place, so you can quickly leverage the best tools for your needs.
All in One AI Platform
Go beyond AI Chat, with Search, Notes, Image Generation, and more.
Cost Savings
Access latest AI models and tools at a fraction of the cost.
Get Sh*t Done
Speed up your work with productivity, work and creative assistants.
Constant Updates
Receive constant updates with new features and improvements to enhance your experience.
Features
Selection of Leading AI Models
Access multiple advanced AI models in one place - featuring Gemini-2.5 Pro, Claude 4.5 Sonnet, GPT 5, and more to tackle any tasks

Speed run your documents
Upload documents to your Zemith library and transform them with AI-powered chat, podcast generation, summaries, and more
Transform Your Writing Process
Elevate your notes and documents with AI-powered assistance that helps you write faster, better, and with less effort
Unleash Your Visual Creativity
Transform ideas into stunning visuals with powerful AI image generation and editing tools that bring your creative vision to life

Accelerate Your Development Workflow
Boost productivity with an AI coding companion that helps you write, debug, and optimize code across multiple programming languages
Powerful Tools for Everyday Excellence
Streamline your workflow with our collection of specialized AI tools designed to solve common challenges and boost your productivity

Live Mode for Real Time Conversations
Speak naturally, share your screen and chat in realtime with AI
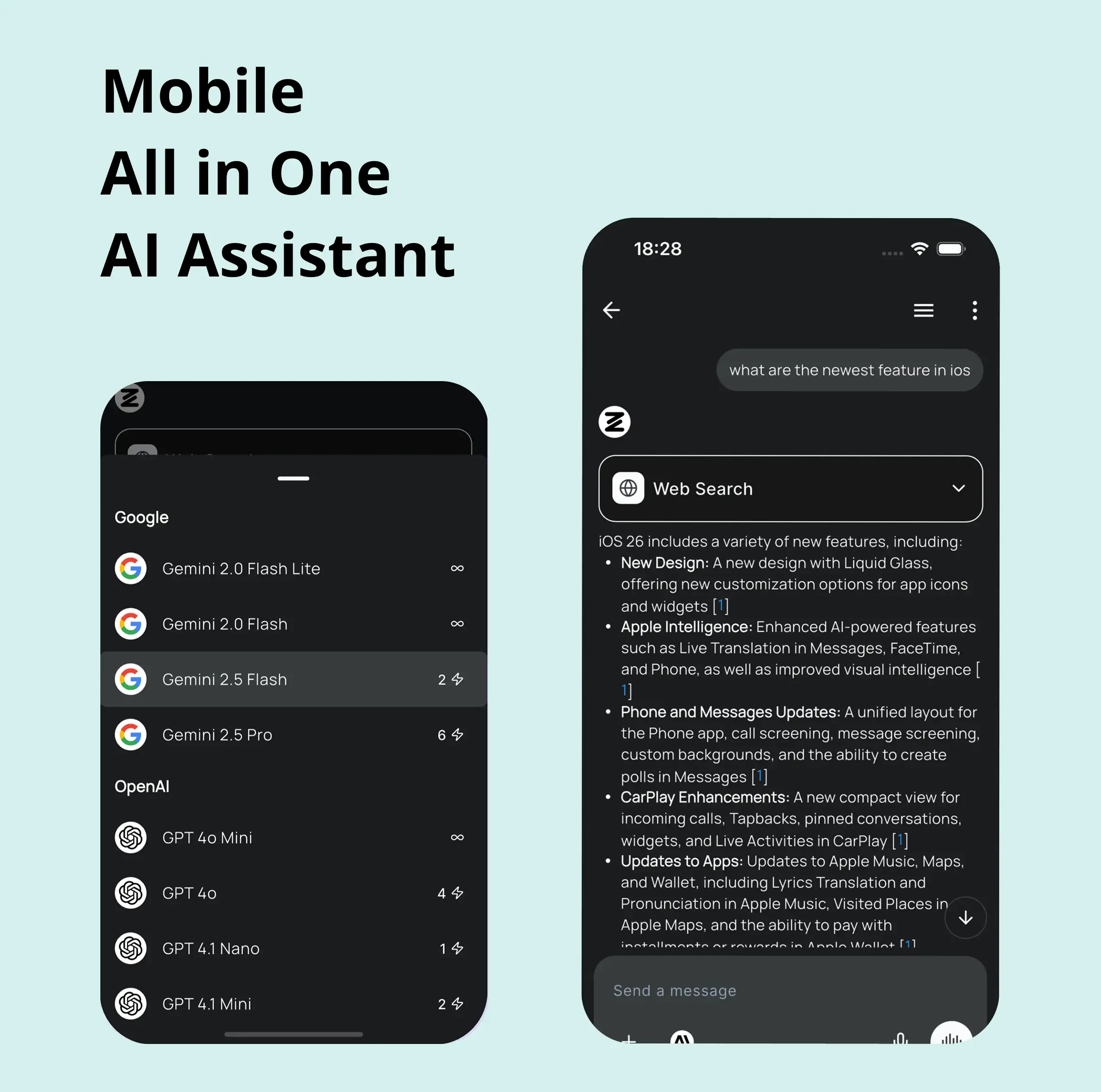
AI in your pocket
Experience the full power of Zemith AI platform wherever you go. Chat with AI, generate content, and boost your productivity from your mobile device.
Deeply Integrated with Top AI Models
Beyond basic AI chat - deeply integrated tools and productivity-focused OS for maximum efficiency
Straightforward, affordable pricing
Save hours of work and research
Affordable plan for power users
Plus
- 10000 Credits Monthly
- Access to plus features
- Access to Plus Models
- Access to tools such as web search, canvas usage, deep research tool
- Access to Creative Features
- Access to Documents Library Features
- Upload up to 50 sources per library folder
- Access to Custom System Prompt
- Access to FocusOS up to 15 tabs
- Unlimited model usage for Gemini 2.5 Flash Lite
- Set Default Model
- Access to Max Mode
- Access to Document to Podcast
- Access to Document to Quiz Generator
- Access to on demand credits
- Access to latest features
Professional
- Everything in Plus, and:
- 21000 Credits Monthly
- Access to Pro Models
- Access to Pro Features
- Access to Video Generation
- Unlimited model usage for GPT 5 Mini
- Access to code interpreter agent
- Access to auto tools
- 10000 Credits Monthly
- Access to plus features
- Access to Plus Models
- Access to tools such as web search, canvas usage, deep research tool
- Access to Creative Features
- Access to Documents Library Features
- Upload up to 50 sources per library folder
- Access to Custom System Prompt
- Access to FocusOS up to 15 tabs
- Unlimited model usage for Gemini 2.5 Flash Lite
- Set Default Model
- Access to Max Mode
- Access to Document to Podcast
- Access to Document to Quiz Generator
- Access to on demand credits
- Access to latest features
- Everything in Plus, and:
- 21000 Credits Monthly
- Access to Pro Models
- Access to Pro Features
- Access to Video Generation
- Unlimited model usage for GPT 5 Mini
- Access to code interpreter agent
- Access to auto tools
What Our Users Say
Great Tool after 2 months usage
simplyzubair
I love the way multiple tools they integrated in one platform. So far it is going in right dorection adding more tools.
Best in Kind!
barefootmedicine
This is another game-change. have used software that kind of offers similar features, but the quality of the data I'm getting back and the sheer speed of the responses is outstanding. I use this app ...
simply awesome
MarianZ
I just tried it - didnt wanna stay with it, because there is so much like that out there. But it convinced me, because: - the discord-channel is very response and fast - the number of models are quite...
A Surprisingly Comprehensive and Engaging Experience
bruno.battocletti
Zemith is not just another app; it's a surprisingly comprehensive platform that feels like a toolbox filled with unexpected delights. From the moment you launch it, you're greeted with a clean and int...
Great for Document Analysis
yerch82
Just works. Simple to use and great for working with documents and make summaries. Money well spend in my opinion.
Great AI site with lots of features and accessible llm's
sumore
what I find most useful in this site is the organization of the features. it's better that all the other site I have so far and even better than chatgpt themselves.
Excellent Tool
AlphaLeaf
Zemith claims to be an all-in-one platform, and after using it, I can confirm that it lives up to that claim. It not only has all the necessary functions, but the UI is also well-designed and very eas...
A well-rounded platform with solid LLMs, extra functionality
SlothMachine
Hey team Zemith! First off: I don't often write these reviews. I should do better, especially with tools that really put their heart and soul into their platform.
This is the best tool I've ever used. Updates are made almost daily, and the feedback process is very fast.
reu0691
This is the best AI tool I've used so far. Updates are made almost daily, and the feedback process is incredibly fast. Just looking at the changelogs, you can see how consistently the developers have ...
